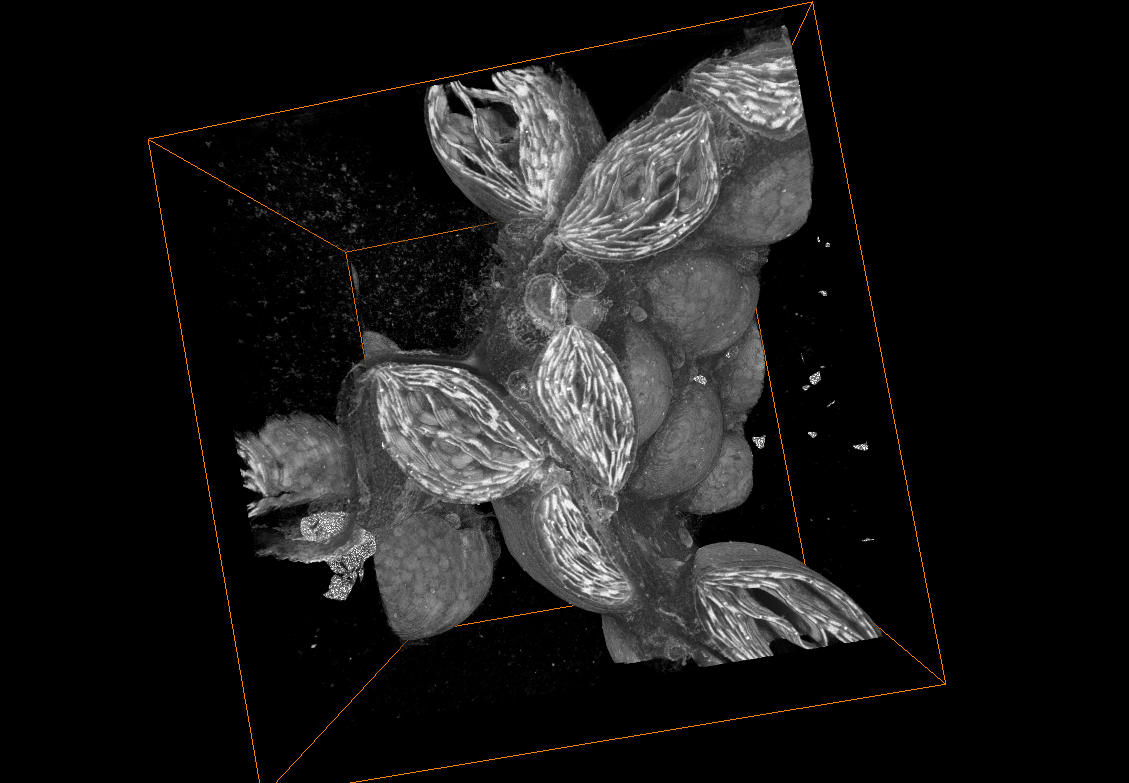
Volume SEM
The volume electron microscopy techniques are well suited to studying complex cellular structures and tissues, as well as nanomaterials. vEM was recently highlighted as one of the ‘seven technologies to watch in 2023’ by Eisenstein (external pageNaturecall_made).
vEM comprises imaging techniques based on transmission electron microscopy (TEM, described in details here) and scanning electron microscopy (SEM). The common principle of these techniques is the acquisition of image series which ultimately are combined into a digital 3D representation of the sample.
vSEM techniques available at ScopeM
Serial Block-Face Scanning Electron Microscopy (SBF-SEM)
This technique involves sequentially removing thin sections from a block of tissue or material using a microtome inside the SEM chamber. After each section is cut, the newly exposed surface is imaged using the SEM. The process is repeated for multiple sections, allowing the reconstruction of a 3D volume of the sample.
ScopeM currently operates a Gatan 3View installed in a TFS Quanta 250. In December 2023, a Gemini SEM 560, equipped with a Zeiss Volutome will be installed.
Contact: Miriam Lucas
Focused Ion Beam Scanning Electron Microscopy (FIB-SEM)
In FIB-SEM, a focused ion beam is used to mill away thin layers from a sample surface. After each milling step, the newly exposed surface is imaged using the SEM. The process is iteratively repeated to obtain a 3D volume. While for SBF-SEM the entire sample surface has to be exposed, FIB-SEM allows a targeted reconstruction of only a specific sub-volume of the specimen. For further information on additional applications of FIB-SEM, please see <<page FIB-SEM>>.
Instruments at ScopeM suited for volume acquisition are TFS Helios 600i and TFS Helios 5 UX.
Contact: Anne Greet Bittermann or Miriam Lucas
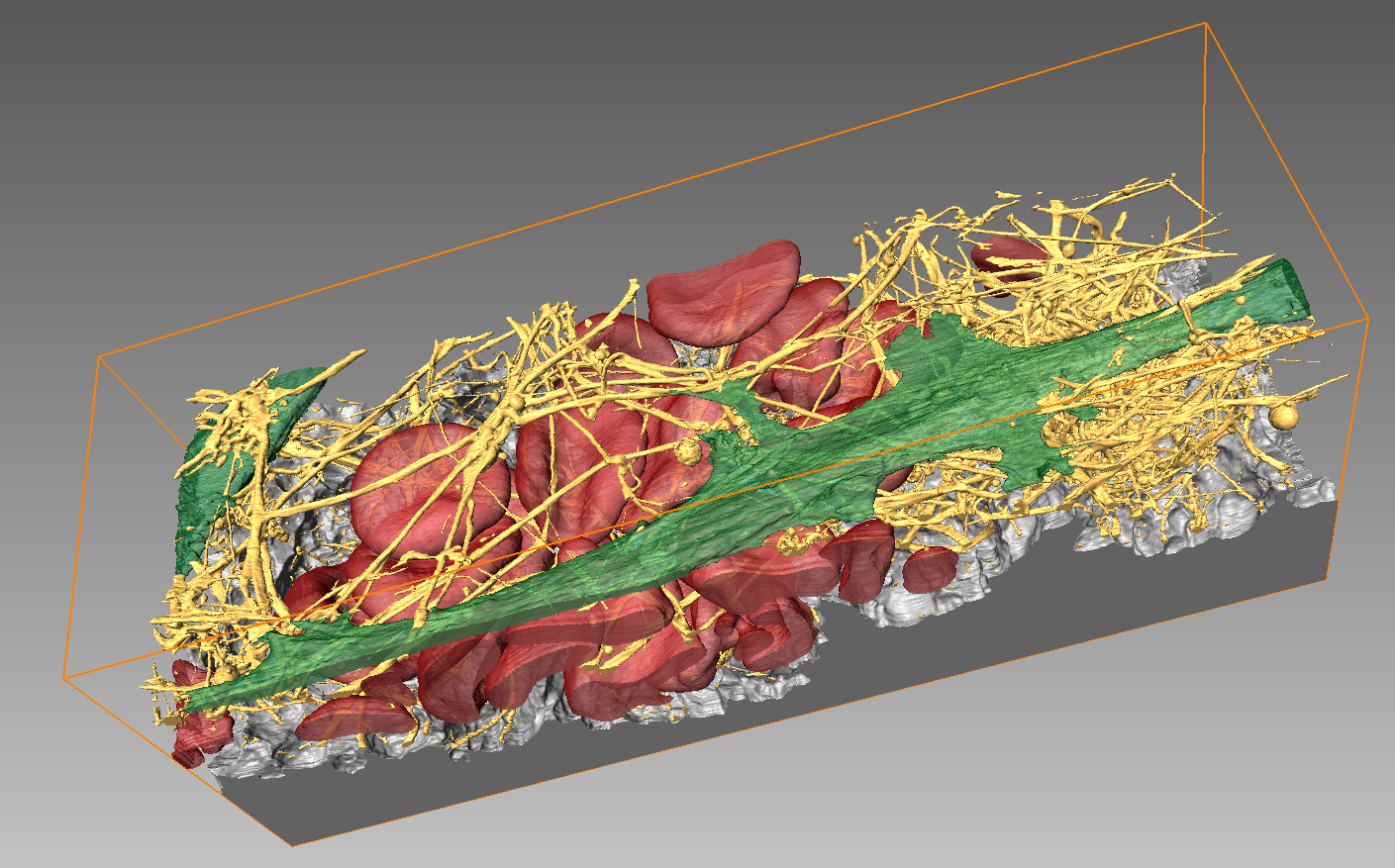
Array Tomography
For array tomography serial sections, so-called arrays, of resin-embedded samples are loaded onto a conductive support suitable for SEM imaging. Selected areas of these consecutive sections are imaged in SEM (see <<large area scanning SEM>>) and the resulting image stacks are combined to form 3D volume data. While SBF-SEM and FIB-SEM are destructive methods, serial sections for AT can be stored and reinvestigated. ScopeM’s Zeiss Merlin is equipped with an Atlas scan system which enables automated image acquisition for AT.
Contact: Falk Lucas or Miriam Lucas