Cryo-EM
Here are the ScopeM sample preparation tools and or .
Moreover, we offer on-the-fly processing using Relion or cryoSPARC-Live during data collection. Furthermore, we offer High Performance Computing (HPC) powered processing pipelines such as cryoSPARC instances in collaboration with ETH Cluster team.
For further information about the cryoSPARC instances or training in image processing workflow packages like Relion or cryoSPARC please contact .
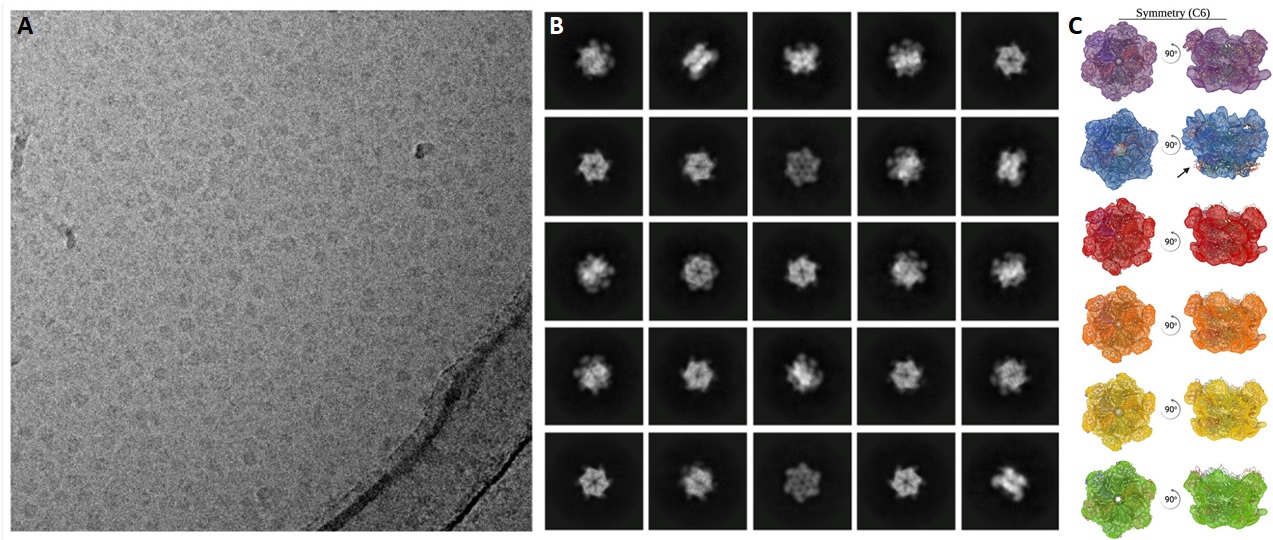
Selected exemplary cryo-EM projects performed at ScopeM are shown below.
Biological macromolecules such as protein complexes or protein-RNA complexes perform various functions in the cell. Near-atomic resolution structures of these molecular machines can be determined using single particle analysis (SPA) technique of cryoEM.
Very large data sets (typically TB) are obtained from thin layers of vitrified specimen on cryo-EM grids. SPA pipelines allow selection of up to millions of particle images depicted in all possible orientations or views. The individual particle images are aligned and averaged in their different views to obtain 2D class-averages of each such orientation with improved signal to noise ratio. Data sets with good angular distribution (comprising many different views) of particles allow reconstruction of 3D volumes. Atomic models can be fitted in such EM maps for further interpretation. For further details see this publication.
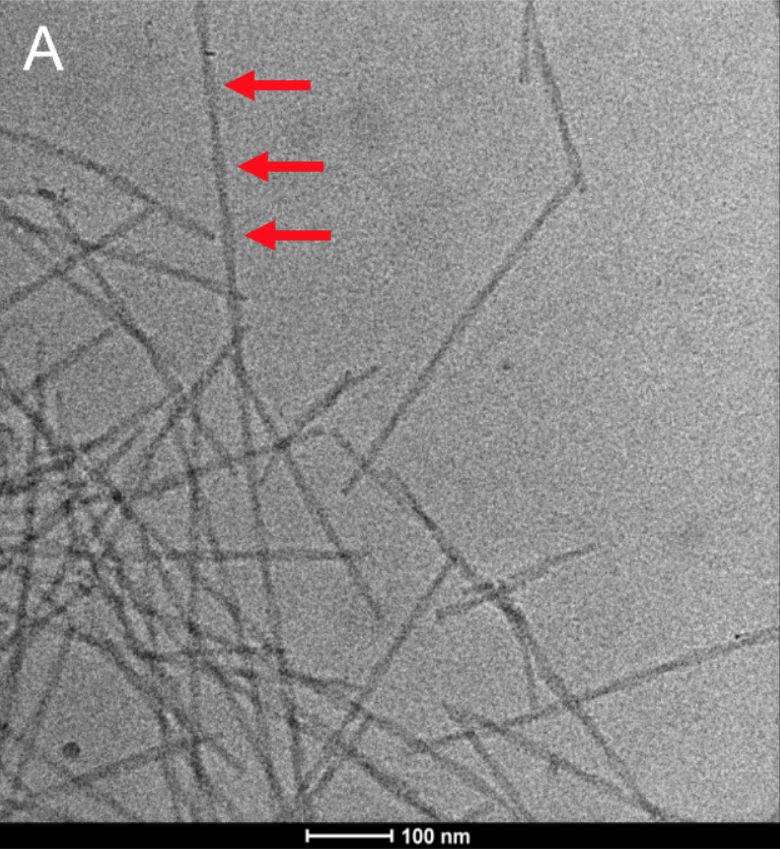
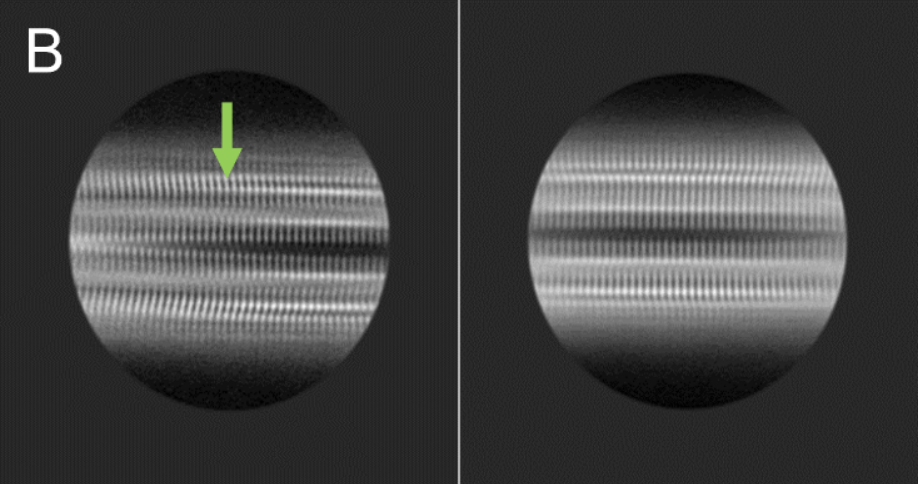
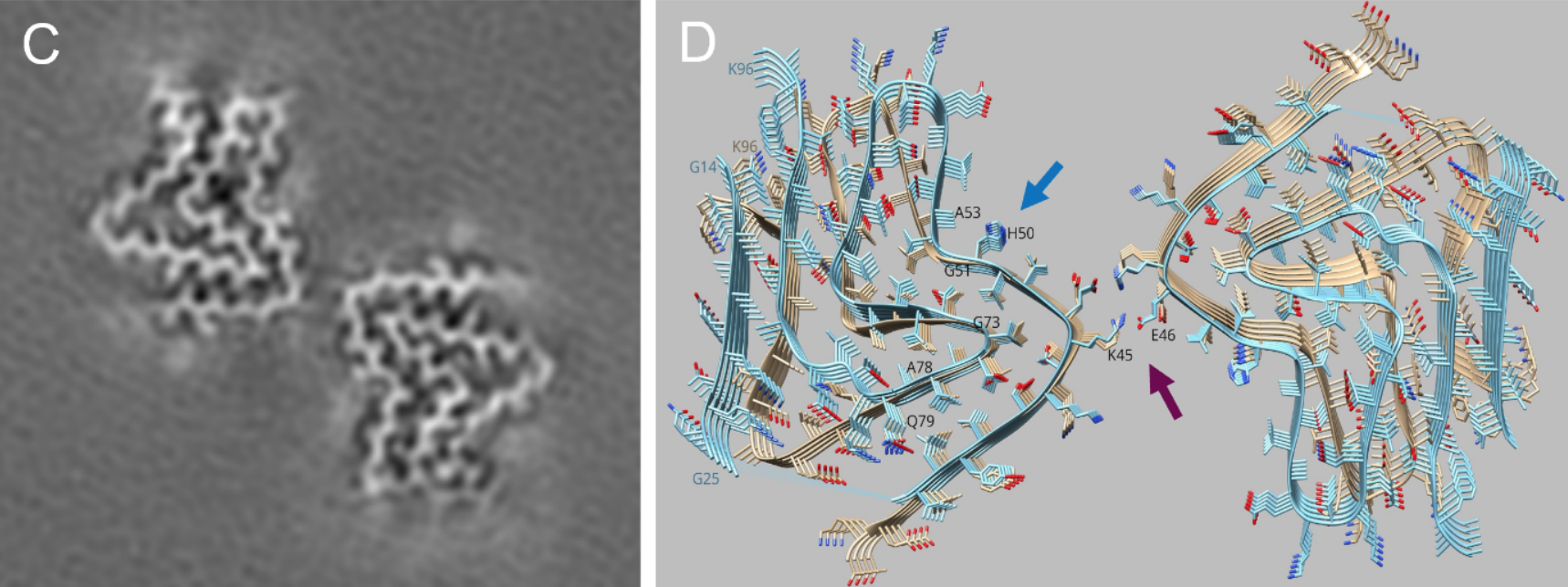
Many biological specimens comprise helical symmetry incl. some viruses (Tobacco Mosaic Virus or TMV), bacterial flagella, DNA, cytoskeleton components such actin or myosin filaments, microtubules etc. Very large data sets (typically TB) are obtained from thin layers of vitrified helical specimen on cryo-EM grids.
Like Single Particle Analysis (SPA), segments of the filaments are selected to obtain 2D class-averages of different sections of the filaments after aligning and averaging them to improve signal to noise ratio. Helical Reconstruction involves symmetry operation of translation and rotation in a screw like fashion to produce the helix. The obtained EM map can be used to fit atomic models for further scientific interpretation.
Helical Reconstruction technique was successfully applied to Parkinson's disease relevant α-synuclein fibrils as shown in Figure below. See also the ScopeM Newsletter.
High Resolution Transmission Electron Microscopy imaging can deliver atomic resolution structures of beam-insensitive or hard materials. However, to achieve such high resolution, it also requires adequate electron dose. Biological samples and soft materials cannot withstand such a high dose, which would "fry" them instantaneously.
Several technological achievements allow successful imaging of such samples using cryo-EM incl. vitrification of the specimen and keeping it under cryogenic temperatures in dedicated cryo TEM Microscopes such as Titan Krios. Moreover, data collection is performed under low dose conditions (typically 30-80 e/A2) on direct electron detectors. Figure below shows cubosome sample imaged on a TFS Tian Krios and Gatan`s K3 detector. Such fine structural details preserved in near-native state are not accessible in images taken under room temperature for these samples.